Chemistry and Biochemistry
Ryno Lab
Research in the Ryno Lab
The Ryno Lab explores new targets that can be utilized in small molecule antibiotic development by activating signaling pathways — interconnected networks of molecular information that dictate cellular activity and health.
View Associate Professor Lisa Ryno's faculty bio and see more about these projects on the Ryno Lab website.
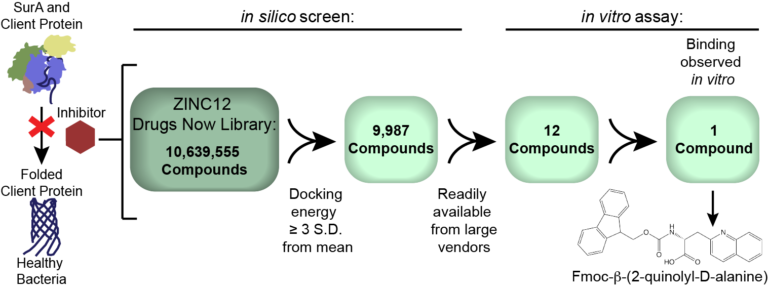
We use computational tools and biochemical assays to discover small molecule inhibitors for the E. coli chaperone SurA that may be useful in the development of new antibiotics.
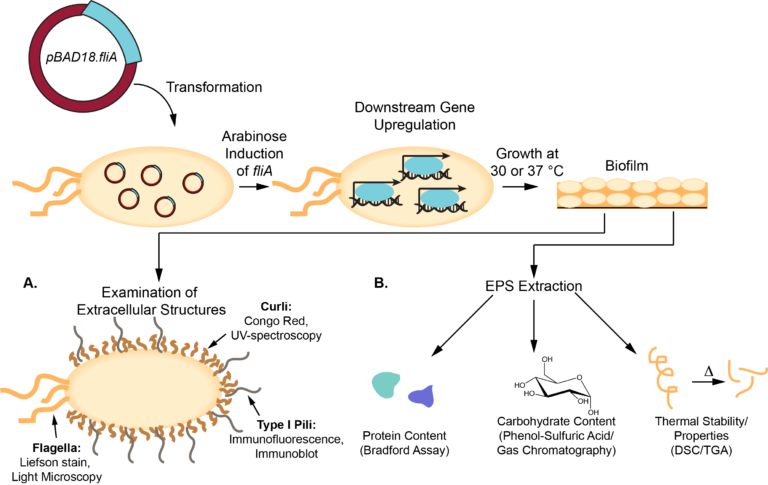
We study the influence of overexpressing stress-responsive transcription factors on the formation and composition of E. coli biofilms. Transcription factors are proteins that can dramatically change the global transcriptome and proteome by changing the expression of several genes simultaneously. Our first transcription factor of interest is fliA, which influences cell motility and chemotaxis. We use colorimetric biochemical assays to determine the concentration of biomolecules like proteins, carbohydrates, lipids and extracellular DNA in the extracellular polymeric substance (the non-cell, viscous matrix that surrounds biofilm cells). We also use Oberlin’s state-of-the-art confocal laser scanning microscope and scanning electron microscope to visualize the influence of these transcription factors on the three-dimensional composition of biofilm.
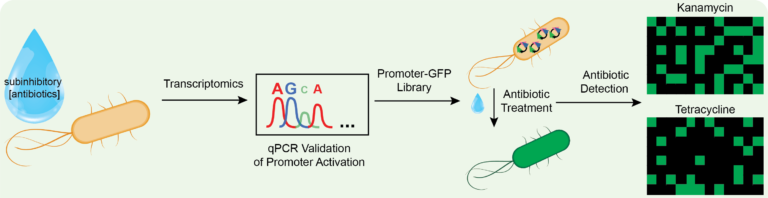
Antibiotic-resistant bacteria and genes can arise from the over-exposure of non-resistant bacteria to antibiotics, including exposure to low concentrations of antibiotics over an extended period. We are studying the use of stress-responsive signaling pathways, pathways that are among the first cellular actions that a bacterium employs when faced with antibiotics or other environmental stressors, as reporters for the detection of low concentrations of antibiotics. We are using transcriptomics to determine those pathways most sensitive to low concentrations of different classes of antibiotics. We are developing a green fluorescent protein (GFP) library that will provide a fingerprint identification for class and concentration of small molecule stressors.